Resistance challenges in veterinary medicine: Who's to blame for "superbugs" and how do we deal with them (Proceedings)
This presentation attempts to summarize some of the major concerns in resistance development along with key articles explaining relevance, epidemiology, and prevalence. It is not intended to be an exhaustive review of the literature and the interested practitioner should use the cited literature herein as a basis for continued, extended reading.
This presentation attempts to summarize some of the major concerns in resistance development along with key articles explaining relevance, epidemiology, and prevalence. It is not intended to be an exhaustive review of the literature and the interested practitioner should use the cited literature herein as a basis for continued, extended reading.
I. So where do these things come from? Here is the basic questions.
• Do resistant organisms develop from spontaneous mutations in your patient (or a population of patients, such as in some food animal applications) during antimicrobial use and then proliferate within the favorable climate of antimicrobial selection pressure?
• Or, are they already present at a low prevalence level and then proliferate in the new environmental "rules" imposed by the presence of antimicrobials (clonal dissemination and selection)?
My impression from the literature and sitting through and participating in meetings, debates, and outright arguments is that dissemination of resistant bacterial clones is a primary driver in what we are seeing in human and veterinary medicine. Spontaneous mutations can and do occur, but the rapid changes in resistance over broad areas, and also the similarities between isolates suggests that the spread of clones is a primary driver.
Another very basic concept is that selection for a resistant pathogen or bacteria may be due to an entirely different selection pressure than the antimicrobial in which we happen to be interested. Multiple-drug resistance mechanisms allow co-selection for resistance traits; and, it doesn't even have to be an antimicrobial in the way we typically think of them. Co-selection by environmental disinfectants can co-select for antimicrobial resistance, as demonstrated for pine oil for E. coli, and triclosan for Pseudomonas aeruginosa. The presence of pathogens such as Vancomycin-Resistant Enterococci (VRE), Pseudomonas, and Methicillin-Resistant Staphylococcus aureus (MRSA) on surfaces, pagers, and stethoscopes has been well documented in human studies.
We don't cause the original spontaneous mutations. But, once these mutations take hold in an environment, we are responsible for aiding in selection and spread. As Pogo said, "We have met the enemy and he is us".
II. What are the challenges on the human side of medicine?
Hospital acquired infections. One publication gives us a quick look into the challenges in human hospitals. These data are from a Centers for Disease Control and Prevention (CDC) summary. The objective was to describe the frequency of selected antimicrobial resistance patterns among pathogens causing device-associated and procedure-associated healthcare-associated infections (HAIs) reported by hospitals in the National Healthcare Safety Network (NHSN). Data were collected on HAIs reported to the Patient Safety Component of the NHSN between January, 2006 and October, 2007. These HAIs included central line-associated bloodstream infections, catheter-associated urinary tract infections, ventilator-associated pneumonia, and surgical site infections. Overall, 463 hospitals reported 1 or more HAIs: 412 (89%) were general acute care hospitals, and 309 (67%) had 200-1,000 beds. There were 28,502 HAIs reported among 25,384 patients. The 10 most common pathogens accounting for 84% of reported HAIs were...
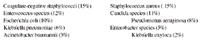
As many as 16% of all HAIs in this report were associated with the following multidrug-resistant pathogens.
• Methicillin-resistant Staph. aureus (8% of HAIs),
• Vancomycin-resistant Enterococcus faecium (4%),
• Carbapenem-resistant Pseudomonas aeruginosa (2%),
• Extended-spectrum cephalosporin-resistant Klebsiella pneumoniae (1%),
• Extended-spectrum cephalosporin-resistant E. coli (0.5%),
• Carbapenem-resistant A. baumannii, K. pneumoniae, K. oxytoca, and E. coli (0.5%).
Challenges in human resistance dissemination: Some examples
Extended-spectrum beta-lactamases: ESBLs were first documented in North America in the middle 1980's in outbreak strains of Klebsiella pneumonia, with a wide variety of genetic families documented since that time. Now, the specter of widely disseminated carbapenem resistance is becoming apparent. The realities of clonal dissemination of this resistance have been documented in the United States, for example recently with a Klebsiella pneumoniae clone in California.
Streptococcus pneumoniae: Changes in macrolide resistance incidence related to target mutation encoded by erm(B) and/or a drug efflux pump encoded by mef(A) have been documented in community-acquired pneumonia in the United States. And, penicillin-resistant Streptococcus pneumoniae has garnered enough attention that a PubMed search on this term retrieves 4277 articles. Even S. pneumoniae resistance to ciprofloxacin has been documented along with increasing prescriptions in Canada from 1997 to 2006.
Methicillin-Resistant Staph aureus (MRSA): Methicillin is no longer commercially available in the United States. It would more accurately be considered "oxacillin resistant", as the concern is for resistance to the beta-lactam-resistant class of penicillins. When resistance is documented to this class, the organism is considered to be resistant to all beta-lactams, including the cephalosporins.
Vancomycin-intermediate and Vancomycin-Resistant Staph aureus: Vancomycin, a glycopeptide, has been a major component of therapy for MRSA. However, emergence of intermediate and resistant MRSA isolates have been a problem well documented over the last decade.
Vancomycin-Resistant Enterococci (VRE): Enterococci (e.g., faecalis and faecium) have emerged as major Gram (+) pathogens in human medicine. As for MRSA, the glycopeptides are critical in therapy of these pathogens. The increasing prevalence of VRE has lead to great concerns for surgical cases with infection, and this concern is heightened as linezolid resistance is also being documented for VRE isolates.
In addition to these selected examples, world-wide concerns about resistance in diseases such as Malaria and Tuberculosis are contributing to anxiety about potentially uncontrollable epidemics.
III. Resistance challenges in veterinary medicine (including zoonotic concerns):
Weese has published an excellent review of antimicrobial resistance issues in companion animals (2008). The primary organisms addressed in this review are 1) Staphylococcus aureus and Staphylococcus pseudintermedius: both methicillin susceptible and resistant, 2) Enterococci: Enterocococcus faecium and Entercoccus faecalis, 3) Streptococci: Strep. zooepidemicus and Strep. Equi in horses, Strep. canis, 3) Escherichia coli, 4) Salmonella spp., and 5) Pseudomonas spp.
Within the limits of this presentation, MRSA will be focused on in order to highlight issues of zoonotic interactions.
Methicillin-Resistant Staph aureus (MRSA): a 2008 review article has summarized literature on animal occurrence, including cattle, dogs, cats, sheep, chickens, horses, rabbits, seals, and psittacine birds. Significant research has been conducted evaluating the potential for exchange of isolates between people and their pets.
Kottler, et al., evaluated the prevalence of MRSA in people and pets in the same household. The sample consisted of one human nasal swab and one dog or cat nasal and fecal swab from 586 households. Staph aureus was classified as methicillin resistant (MRSA) or susceptible (MSSA). Pulsed-field gel electrophoresis (PFGE) and spa-typing were used to characterize the relatedness of S. aureus and MRSA between pets and humans. There was no difference in MRSA prevalence in households with human healthcare workers, veterinary healthcare workers, or without healthcare workers. The following table displaying prevalence of MSSA and MRSA in humans and pets is adapted from the publication.

In 4 of the 586 households (0.7%), the MRSA found in humans was the same strain as that found in the pet.
Faires, et al., evaluated the prevalence of concurrent infection in households where either a person or pet had a diagnosed MRSA colonization. In part 1 of the study, 22 households were identified as having an MRSA infection in a pet (19 dogs and 3 cats). In these households, 10 of 56 humans (17.9%) were also colonized with MRSA. In part 2 of the study, 8 households were identified where humans had MRSA cultures from dermal abscesses. In only 1of these households was MRSA also isolated from a pet. In almost all cases of co-colonization or infection, the isolates were indistinguishable by PFGE.
O'Mahony, et al., evaluated MRSA isolates from dogs, horses, a cat, a rabbit, and a seal in Ireland along with isolates from 10 caregivers. The PFGE results for the equine MRSA isolates were indistinguishable from the results for those isolates originating from the caregivers for the horses.
Several studies have evaluated risk factors for infection with MRSA in companion animals. Faires, et al., evaluated risk factors for 40 MRSA infected dogs compared with 80 MSSA infected dogs. The highest prevalence of both infections was in ears and skin. The statistically significant risk factors for MRSA infection as compared to MSSA infection included the use of any antimicrobial prior to diagnosis (odds ratio 2.84), use of fluoroquinolones (OR 3.58), use of β-lactams (OR 3.58), or intravenous catheterization (OR 3.72).
A retrospective study in horses in Canadian and American referral hospitals evaluated MRSA infections in 115 horses. The infections originated both in the referral hospitals and in the community, with the frequency of both being approximately equal. Community acquired infections were significantly associated with previous hospitalization and previous gentamicin therapy. Hospital-acquired MRSA infections were significantly associated with infected incision sites.
Increasing attention in the literature has been paid to MRSA in swine and potential zoonotic concerns. There is extensive literature on types and occurrence of MRSA in farm workers. While swine workers and veterinarians have been demonstrated to have nasal carriage of the MRSA type found in swine herds, epidemiological studies suggest that colonization is primarily limited to those working with the swine and further transmission is limited to familial communities of these exposed workers. In the U.S., the human community-acquired outbreak strains are different from animal strains. In the Netherlands, a new type of MRSA (ST 398) is epidemiologically associated with pig and cattle farmers and is said to be > 20% of carriage in humans. However, this type has not been associated with significant human disease. MRSA has also been identified in bovine mastitis isolates.
MRSA is an example of a resistant organism (which may also be multi-drug resistant) that brings the issue of treating our veterinary patients together with concerns about the effect of this pathogen's presence on our clients. There are no free lunches, as pathogens which have developed resistance to one main line of therapy will likely also develop resistance to the next great thing in therapy. Therapeutic strategies for these resistant pathogens are the subject of the next presentation.